PhD Project
Genetic diversity in non-native conifer UK plantations and their offspring managed under the Continuous Cover Forestry approach

Even-aged plantations of non-native conifer species and clear-cutting silvicultural practices are widely used in UK forestry to maximize timber harvest yield. However, increasing concern for the adaptability of forests to future and current environmental shifts is leading to changes in forestry management. Continuous Cover Forestry (CCF) is an approach to managing forests based on the development of diverse stand structures composed of a mixture of species and encouraging natural regeneration while keeping a timber production aim. While the transition to this type of woodland is a well-defined silvicultural approach, there is a current paucity of studies on how the transmission of the gene pool from canopy trees to the next generation may affect the genetic diversity of future forest stands. The canopy that was planted may not show a large genetic diversity resulting in a poor gene pool being transmitted to the offspring resulting in forests lacking the ability to face potential environmental changes. To estimate how genetically diverse these plantations are, we will look at two species widely used for timber production in the UK and will genotype individuals from different strata to estimate how the gene pool is transferred. We will use available Single Nucleotide Polymorphisms (SNPs) datasets described previously in the literature for Pseudotsuga menziesii (Douglas-fir ) and de novo SNP discovery in Thuja plicata (Western red cedar). Douglas-fir and Western red cedar SNP databases were filtered, and 96 SNPs were selected for further genotyping analysis. A SNPs genotyping approach was developed in a high-throughput microfluidic PCR system (Biomark system, Fluidigm®). We’ll validate the method by genotyping a subsample of Douglas-fir individuals using IPEX Gold massarray platform together with individuals from known provenance trials. The genotypic comparison between generations will shed light upon the vulnerability of such populations leading to the recommendation of comprehensive and effective genetic conservation strategies when needed.
MSc Research Project
RADdesigner: a workflow to select the optimal sequencing methodology in genotyping experiments on woody plant species

Polymorphism detection in woody plant species is important in phylogenetics and an essential stage in plant breeding programs. Genotyping has benefitted from the development ofnext-generation sequencing (NGS) techniques. Methodologies based upon single and double-digested DNA sequencing (RADseq and ddRADseq) allow the detection ofpolymorphisms on thousands of loci in non-model organisms for which a reference genome is not available, such as many woody plant species. However, experiments using these methodologies are not safe from technical errors, and require careful designs in order to reduce library preparation and sequencing costs at the same time unbiased results are obtained. In order to avoid undesired effects oftechnical errors on genotyping, it is advisable to incorporate error quantification through pilot sequencing of technical replicates, along with filtering strategies and parameter tuning, so the technical conditions of the whole experiment can be properly defined a priori. However, these kinds ofapproaches to account for technical errors have been frequently disregarded in the literature. Here, we present RADDESIGNER, a workflow that combines pilot sequencing ofreplicate samples with a bioinformatic pipeline to select the most suitable methodology by quantifying genotyping errors, to optimize polymorphism detection parameters, and to perform post-filtering of the results produced by the commonly used RADseq bioinformatic pipeline IpyRAD. The workflow was validated using a pilot study that analyzed a subset of replicate samples ofQuercus suber, Q. ilex, and their hybrids.
BSc Research Project
Genetic differentiation of Quercus faginea and Q. pubescens populations across the Pre-Pyrenees
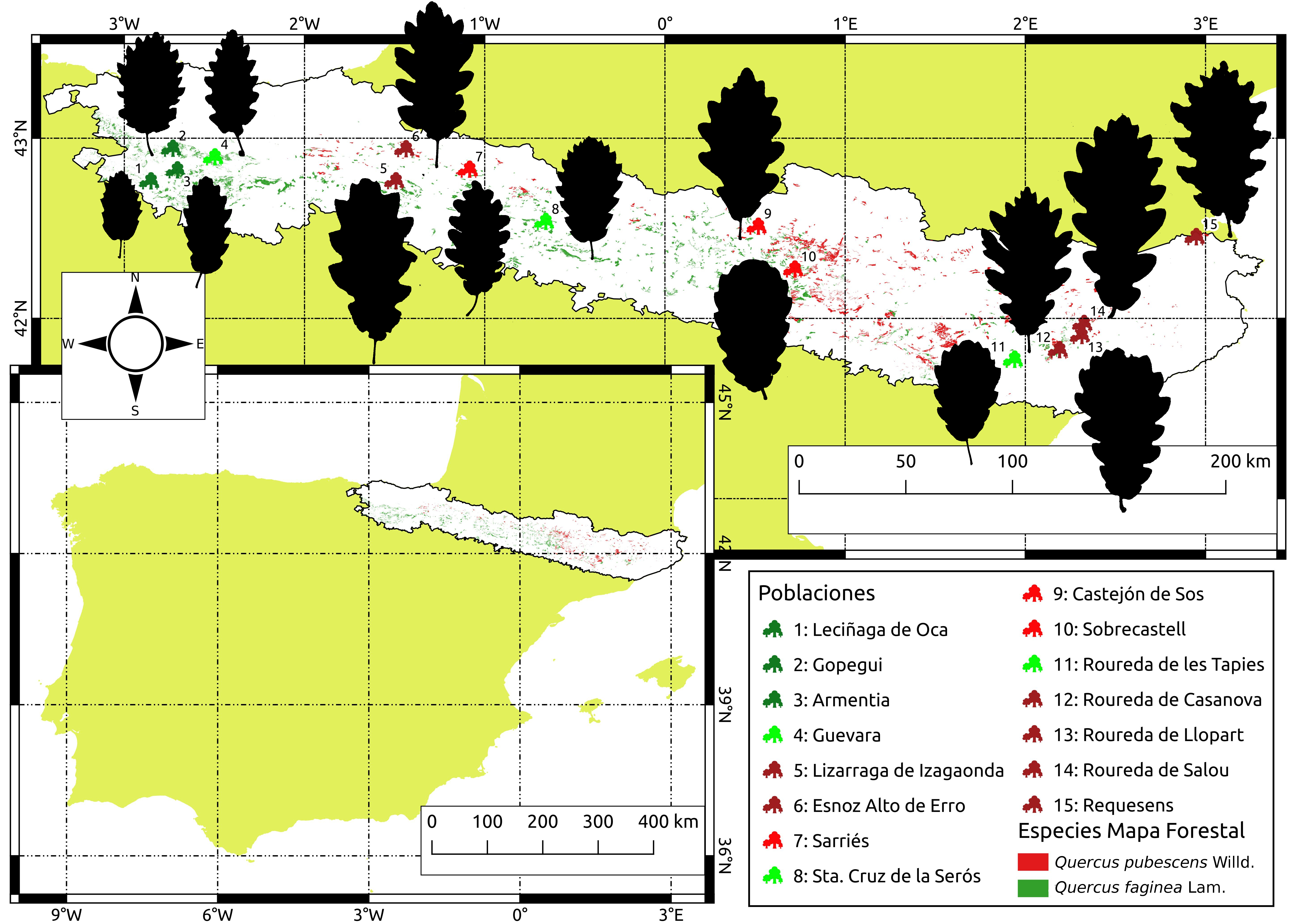
The European white oaks (Genus Quercus subgen. Quercus L.) is a group of species that form a singameon, i.e. a group of species which hybridize and act as a biological species, producing fertile individuals. The complex of species Quercus faginea Lam. - Quercus pubescens Willd. is part of that singameón. In the pre-pyrenean area, where these species may occur forming mixed stands in sympatry, the taxonomic determination of individuals is of great difficulty. In fact, the huge range of phenotypic characters which are presented by these individuals throughout this area suggests that hybridization processes are taking place. Along this project, eight individuals have been investigated in each of the 15 populations sampled within the pre-pyrenean range, with the aim of perform a genetic characterization. There were used 105 microsatellites (EST-SSRs), and methodologies implemented in the Structure and Tess3R software have been used to determine the most probable number of genetic groups in the populations studied, as well as the allocation of individuals to these genetic groups. The results of the genetic analysis show diferences between the methodologies used. The software STRUCTURE show the existence of two genetic groups, corresponding to the complex of species Quercus faginea Lam. - Quercus pubescens Willd, and a considerable number of individuals showing high genetic admixture. On the other hand, the methodology that implements the software TESS3R shows the existence of a single genetic group.